Ntobeko Dube and Idah Sithole-Niang*
Department of Biotechnology and Biochemistry, University of Zimbabwe, Zimbabwe
*Corresponding Author:Idah Sithole-Niang, Department of Biotechnology and Biochemistry, University of Zimbabwe, Zimbabwe.
Received: February 08, 2021; Published: February 27, 2021
Endophytes are microbes found within live plant tissues and are known to be reservoirs of bioactive compounds that promote plant growth and antimicrobial activity. This makes them important, but are understudied. In this study endophytes from Diplorhynchus condylocarpon plant were characterized at molecular level and their potential to produce bioactive compounds with antimicrobial activity was explored. DNA extraction was carried out on twenty-four endophytes isolated from leaves and twigs of Diplorhynchus condylocarpon. To confirm the nature of these endophytes at kingdom level the 16S rRNA gene was amplified using the Polymerase chain reaction and all indicated to be prokaryotes in nature. Nine 16S rRNA amplicons out of twenty-four were sequenced and species were identified as Izhakiella capsodis, Escherichia fergusonii, Shigella dysenteriae, Xernorhabdus szentirmaii, Providencia rettgeri, Dickeya zeae and Escherichia albertii. Agar well diffusion was used to test for antimicrobial activity of twenty-four endophytes. All twenty-four endophytes exhibited antimicrobial activity against E. coli and S. aureus, with wide range of zones ranging from 19 to 65 mm. Using KEGG database it was discovered that sequenced species have potential to produce antibiotics such as fosfomycin and streptomycin. Secondary agar well diffusion was done to compare potency of endophytes extracts with ampicillin, a known antibiotic. Endophytic extracts were observed to be more effective than ampicillin with highest zones of inhibition for extracts and ampicillin as 54 and 36 respectively. In conclusion this study showed that endophytes from Diplorhynchus condylocarpon are reservoirs of bioactive compounds with antimicrobial effect. Also, that the 16S rRNA gene can be used to identify species.
Keywords: Diplorhynchus condylocarpon; Endophytes; Molecular Characterization; Bioactive Compounds; Antimicrobial Assay; 16S rRNA
Use of traditional plants for medicinal purposes has been one of the most affordable and easy access to treatment of poor communities in Zimbabwe and there is a long history of use of plants for medicinal purposes. About 10% of all species growing in Zimbabwe have medicinal properties. Within 10% shrubs and trees taking 38% each, herbs at 21% and climbers at 3%. Diplorhynchus condylocarpon also known as Mutowa in Shona is a plant found in Zimbabwe and other tropical, southern African countries. It can grow either as a shrub or a tree and has been used for medicinal purposes.
With an increase in drug resistance which has become a global health issue, new methods to solving this crisis and exploring medicinal properties of endophytes have been adopted by many countries. Endophytes are microbes that reside inside living plant tissues. Endophytes are phylogenetically rich, hyper-diverse species, ecologically important and are under explored species with wide range of genetic and functional diversity. Taxonomic classification of endophytes begins at the kingdom level (Fungi or bacteria). Almost every plant species across the globe is colonized by endophytes [17]. No visible harm has been noticed to be caused by endophytes in a plant they colonize. Relationship between the plant and endophytes is usually mutualistic [1].
Molecular characterization is a technique used to differentiate and identify organisms by characterizing at the molecular level without depending on factors such as environment, physiological state or development of an organism [4]. Molecular techniques involving the use of small ribosomal RNA (rRNA) sequence variability as a biomarker has been used mostly for the past years to characterize bacterial endophytes. There are several advantages associated with using 16S rRNA gene sequence as a biomarker in molecular characterization of bacterial endophytes because, these include its presence in all prokaryotes species, high variation across species and presence of many sequences of 16S rRNA from various organisms are present in the database and this gives reference sequences when identifying species [11].
Despite the identification of endophytes and potential they pose in many biological applications, there is no significant information about endophytes in Africa before 1994. Between 1994 and 2014 Egypt and South Africa were the two leading African countries to have researched on endophytes. In Zimbabwe use of endophytes for medicinal purposes and other biological applications has been approached in recent years [16]. Diplorhynchus condylocarpon also known as mutowa in Shona is a plant found in Zimbabwe and other tropical, southern African countries. Diplorhynchus condylocarpon has been used traditionally to treat several infections such as indigestion, diarrhea, fever, snakebites and infertility. Endophytes from Diplorhynchus condylocarpon are distributed in all parts of the plant therefore characterization of these endophytes is less expensive and can be used in biotechnological processes.
In this study endophytes from Diplorhynchus condylocarpon plant were characterized at molecular level and their potential to produce bioactive compounds with antimicrobial activity was explored. The 16S rRNA sequencing technique is also used for identification of the bacterial species.
The study was carried using endophytes isolated from two Mutowa trees obtained from different geographical locations (Kwekwe and Karoi). The endophytic isolates were procured from the glycerol stocks prepared by Ms Jordanca Kugara. To induce growth, the endophytic isolates were cultured on potato dextrose agar (PDA) and incubated at room temperature for 24 hours. Each colony that grew was sub-cultured in Luria Bertani broth (LB) from Kirkhouse Trust England and incubated at room temperature for 4 days.
Media preparation Agar plates preparationThe media was prepared by dissolving 40 g of PDA powder in 1000 ml distilled water and the mixture was autoclaved for 15 minutes at 121oC. The sterile molten PDA media was poured in sterile petri dishes and left to solidify.
LB broth preparationThe liquid media was prepared by dissolving 11.5 g of LB broth powder in 500 ml of distilled water. The mixture was then autoclaved for 15 minutes and 121oC. The media was allowed to cool before inoculation of endophytic isolates.
Genomic DNA extractionAn aliquot of 500 ul of CTAB buffer was mixed with 500 ul of liquid overnight culture in a 1.5 ml centrifuge tube. The mixture was then incubated at 55°C for 30 minutes. After the incubation the samples were centrifuged for 5 minutes at 5000 rpm and the supernatants were then transferred to new tubes. Equal volume of chloroform was then added to each tube, centrifuged at 5000 rpm for 3 minutes and the supernatants were transferred to new tubes. Subsequently, an equal volume of ice-cold isopropanol was added to each tube and the mixture was incubated for 20 minutes at -20°V. Afterwards, the tubes were centrifuged at 10000 rpm for 10 minutes and the supernatants were discarded. The pellets formed were washed by adding 500 ul of ice-cold 70% ethanol and centrifuged at 10000 rpm for 3 minutes. The supernatant was discarded and the pellets were dried in a speed vac for 10 minutes. An aliquot of 50 ul of TE buffer was added into each tube to dissolve the nucleic acids. The quality of DNA in each sample was evaluated by electrophoresis in 0.8% agarose gel at 100 V for 30 minutes. Afterwards, the agarose gel was visualized under UV light and a picture of the DNA bands was taken.
16S PCR assayThe bacterial 16S rRNA was amplified using the genomic DNA template and 27F (5’- AGAGTTTGATCCTGGCTCAG -3’) forward primers and 1492R and (5’-CGGTACCTTGTTGTTACGACTT-3’) reverse primer. The PCR reactions were carried out with 25 ul master mix containing (5 ul 5 x PCR buffer,2.5 ul 2 mM dNTPs, 1.5 ul 25 mM MgCl2, 3 ul Taq DNA polymerase, 1 ul 27F primer, 1 ul 1492R primer and 1 ul endophytic genomic DNA). The PCR was carried out in a thermo cycler (Applied Biosystems, model 2720) under these conditions, 95°C for 5 minutes for denaturation, 95°C for 45 seconds for further denaturation, 60°C for 40 seconds for annealing of DNA with primers, primer elongation at 72°C for 3 minutes and a final extension for 5 minutes still at 72°C to complete the full cycle. The PCR machine was set to stop after thirty-five cycles. The amplified DNA was then evaluated by electrophoresis in 1% gel at 100 V for 30 minutes. The gel was visualized using UV light and the picture of bands was taken.
16S rRNA gene sequencingThe 16S rRNA gene amplicons were sent to Stellenbosch University in South Africa for sequencing.
Phylogenetic analysisMultiple sequence alignment was done on 16S rRNA gene sequences using Clustal omega program. This program makes use of seeded guide trees and Human Markov Models profile-profile technique in aligning sequences. After aligning sequences this program builds a phylogenetic tree.
Antibacterial assay Extraction of bioactive compounds from endophytesAn aliquot of 200 ul 10% SDS was mixed with 600 ul of liquid 4 days culture in a 2 ml centrifuge tube. The mixture was then mixed and incubated at room temperature for 4 minutes. After incubation samples were centrifuged for 5 minutes at 10000 rpm. Supernatants were transferred to new tubes. Equal volume of acetone was added to each tube to, centrifuged for 3minutes at 5000 rpm. After centrifugation top aqueous layer which contained extracted bioactive compounds was then transferred to different boats of known mass and samples were dried overnight in the laminar hood. Dried extracts were then resuscitated using 120 ul 2% DMSO. The same process was repeated using two other solvents methanol and ethyl acetate. However, for ethyl acetate samples were dried and resuscitated within the same day because ethyl acetate extracts dry fast.
Preparation of tester platesTest microorganisms, E. coli (ATCC 35218) and S. aureus (ATCC 25923) were obtained from University of Zimbabwe department of biochemistry. Overnight cultures of bacterial tester strains were prepared by mixing 1 ml of tester strain and 10 ml of LB broth and incubated at 37oC. The following day OD of the cultures was measured using spectrophotometer and standardized to 1 OD by diluting with LB broth. In the hood a volume of 100 ul of 1 OD bacterial tester strain culture was then spread on top of PDA plates. Plates were then left for 1 hour and then 3 holes were created on the plates using the back of 200 ul pippete tips. In the first hole 10 ul of endophytic extract was added, on the other holes 25 ul and 50 ul volumes of extract were added. Plates were placed in the 37 oC incubator and observed after 24, 48 and 96 hours
Preparation of plates with ampicillin as a controlTester plates were prepared almost the same way as mention above. The only difference is that acetone alone was used as a solvent and in place of 25 ul extract, 10 ul of ampicillin was added. Plates were then incubated and observed after 24 and 48 hours.
Twenty-four bacterial endophytes were isolated from two sets of mutowa tree (Kwekwe and Karoi respectively). 60% of endophytes were isolated from the leaves and 40% from twigs. All endophytes isolates were able to inhibit E. coli and S. aureus in agar well diffusion assay and zones of inhibitions ranged from 22 mm to 65 mm for 50 ul extract (Table 3). Extracts were effective on S. aureus to a greater extent compared to E. coli. This study showed broad antimicrobial activity spectrum against both Gram negative and Gram-positive bacteria. Significant difference in antimicrobial effect was noted between two strains used (S. aureus and E. coli). S. aureus was more susceptible compared to effect on E. coli. This correlates to study carried out by [10] that observed that Gram positive bacteria and more susceptible to antibiotics compared to Gram negative, this is because Gram negative have largely impermeable cell wall. Also [3] who found out that endophytes from Azadirachta indica produced bioactive compound that had antibacterial effect against both gram negative and gram positive bacteria.
From this study it was observed that most acetone extracts had highest zones of inhibition followed by ethyl acetate and low antimicrobial potency was observed in methanol extracts. According to [2] acetone and ethyl acetate are polar aprotic solvents meaning they are capable of extracting both polar and non- polar compounds while methanol is a polar protic solvent that can extracts polar compounds. Difference in yield between acetone and Ethylacetate indicate that acetone is a better solvent. Different volumes of extracts from endophytes were used for the agar well diffusion assay, with the 50 ul showing a greater inhibition compared to the 10ul and 25 ul volumes (Table 1 and 2). Highest zones of inhibition were observed from sample 5, 8, 15, 18, 20, 22. All samples were observed to have broad spectrum against both tester strains with all the three solvents. Nonetheless sample 6, 7, 9, 13, 14 and 21 showed less potency.
| Sample | Acetone | Methanol | Ethyl acetate | ||||||
|---|---|---|---|---|---|---|---|---|---|
10 ul |
25 ul |
50 ul |
10 ul |
25 ul |
50 ul |
10 ul |
25 ul |
50 ul |
|
mm |
mm |
mm |
mm |
mm |
mm |
mm |
Mm |
mm |
|
1-Leaf |
20 |
26 |
29 |
23 |
31 |
36 |
25 |
33 |
50 |
2-Leaf |
28 |
34 |
43 |
25 |
29 |
33 |
23 |
30 |
38 |
3-Twig |
26 |
39 |
33 |
29 |
32 |
43 |
27 |
35 |
45 |
4-Leaf |
30 |
41 |
42 |
10 |
21 |
26 |
22 |
35 |
37 |
5-Leaf |
29 |
34 |
53 |
25 |
26 |
32 |
15 |
19 |
27 |
6-Twig |
33 |
35 |
35 |
22 |
28 |
34 |
-- |
-- |
-- |
7-Twig |
37 |
37 |
38 |
24 |
29 |
41 |
-- |
-- |
-- |
8-Twig |
39 |
32 |
37 |
30 |
32 |
40 |
25 |
25 |
30 |
9-Leaf |
25 |
36 |
55 |
23 |
25 |
39 |
-- |
-- |
-- |
10-Leaf |
36 |
34 |
35 |
19 |
24 |
38 |
27 |
27 |
40 |
11-Twig |
19 |
24 |
35 |
24 |
25 |
45 |
22 |
22 |
30 |
12-Leaf |
37 |
37 |
40 |
23 |
27 |
35 |
21 |
28 |
35 |
13-Twig |
27 |
27 |
38 |
30 |
28 |
32 |
-- |
-- |
-- |
14-Leaf |
39 |
30 |
40 |
27 |
23 |
39 |
21 |
34 |
35 |
15-Twig |
17 |
35 |
33 |
18 |
22 |
31 |
25 |
40 |
51 |
16-Leaf |
35 |
35 |
47 |
21 |
30 |
34 |
35 |
35 |
46 |
17-Twig |
26 |
33 |
39 |
22 |
23 |
35 |
19 |
25 |
37 |
18-Leaf |
24 |
29 |
55 |
21 |
32 |
34 |
28 |
30 |
42 |
19-Leaf |
16 |
23 |
41 |
27 |
32 |
35 |
27 |
33 |
50 |
20-Leaf |
22 |
29 |
48 |
24 |
26 |
30 |
39 |
36 |
52 |
21-Leaf |
19 |
22 |
34 |
30 |
24 |
32 |
-- |
-- |
-- |
22-Leaf |
20 |
32 |
59 |
22 |
23 |
40 |
33 |
44 |
58 |
23-Leaf |
23 |
36 |
37 |
23 |
25 |
40 |
27 |
31 |
31 |
24-Twig |
23 |
43 |
41 |
20 |
19 |
30 |
26 |
30 |
35 |
Table 1: Zones of inhibition observed at day 4 in well diffusion assays using solvent extracts (acetone, methanol an ethyl acetate) from 24 bacterial endophytes at different concentrations. S. aureus was used as tester strain. Zones were measured in millimeters (mm).
| Sample | Acetone | Methanol | Ethyl acetate | ||||||
|---|---|---|---|---|---|---|---|---|---|
10 ul |
25 ul |
50 ul |
10 ul |
25 ul |
50 ul |
10 ul |
25 ul |
50 ul |
|
mm |
mm |
mm |
mm |
mm |
mm |
mm |
Mm |
mm |
|
1-Leaf |
29 |
35 |
33 |
18 |
22 |
26 |
20 |
24 |
35 |
2-Leaf |
19 |
19 |
36 |
15 |
20 |
23 |
31 |
31 |
48 |
3-Twig |
34 |
40 |
45 |
13 |
18 |
26 |
32 |
32 |
45 |
4-Leaf |
15 |
22 |
24 |
15 |
18 |
32 |
20 |
21 |
28 |
5-Leaf |
17 |
21 |
29 |
18 |
21 |
26 |
29 |
38 |
40 |
6-Twig |
16 |
20 |
30 |
20 |
25 |
31 |
21 |
27 |
27 |
7-Twig |
20 |
20 |
38 |
16 |
19 |
31 |
-- |
-- |
-- |
8-Twig |
27 |
28 |
45 |
15 |
20 |
22 |
23 |
37 |
55 |
9-Leaf |
30 |
30 |
36 |
19 |
24 |
44 |
32 |
33 |
46 |
10-Leaf |
25 |
31 |
31 |
-- |
17 |
25 |
33 |
40 |
40 |
11-Twig |
22 |
22 |
24 |
15 |
19 |
27 |
25 |
36 |
44 |
12-Leaf |
17 |
18 |
22 |
20 |
26 |
30 |
25 |
34 |
34 |
13-Twig |
16 |
20 |
22 |
17 |
20 |
25 |
25 |
38 |
41 |
14-Leaf |
18 |
19 |
30 |
17 |
25 |
26 |
20 |
20 |
20 |
15-Twig |
14 |
24 |
30 |
14 |
19 |
26 |
27 |
34 |
40 |
16-Leaf |
19 |
20 |
24 |
19 |
20 |
29 |
23 |
30 |
30 |
17-Twig |
19 |
35 |
35 |
20 |
20 |
31 |
20 |
30 |
38 |
18-Leaf |
18 |
19 |
22 |
21 |
25 |
30 |
21 |
35 |
39 |
19-Leaf |
13 |
22 |
29 |
33 |
33 |
58 |
31 |
31 |
50 |
20-Leaf |
14 |
25 |
26 |
15 |
17 |
22 |
29 |
42 |
65 |
21-Leaf |
29 |
30 |
30 |
22 |
25 |
27 |
-- |
21 |
29 |
22-Leaf |
13 |
18 |
24 |
32 |
32 |
38 |
-- |
17 |
19 |
23-Leaf |
11 |
18 |
23 |
21 |
26 |
30 |
26 |
26 |
46 |
24-Twig |
17 |
21 |
35 |
16 |
19 |
25 |
26 |
27 |
34 |
Table 2: Zones of inhibition observed at day 4 in well diffusion assays using solvent extracts (acetone, methanol an ethyl acetate) from 24 bacterial endophytes at different concentrations. E. coli was used as tester strain. Zones were measured in millimeters (mm).
| Sample | Acetone | Methanol | Ethyl acetate | |||
|---|---|---|---|---|---|---|
S. aureus Inhibition zones/mm |
E. coli Inhibition zones/mm |
S. aureus Inhibition zones/mm |
E. coli Inhibition zones/mm |
S. aureus Inhibition zones/mm |
E. coli Inhibition zones/mm |
|
1-Leaf |
29 |
33 |
36 |
26 |
50 |
35 |
2-Leaf |
40 |
36 |
33 |
23 |
38 |
48 |
3-Twig |
39 |
45 |
43 |
26 |
45 |
45 |
4-Leaf |
42 |
24 |
26 |
32 |
37 |
28 |
5-Leaf |
53 |
29 |
32 |
26 |
27 |
40 |
6-Twig |
35 |
30 |
34 |
31 |
-- |
27 |
7-Twig |
38 |
38 |
41 |
31 |
-- |
-- |
8-Twig |
39 |
45 |
40 |
22 |
30 |
55 |
9-Leaf |
55 |
36 |
39 |
44 |
-- |
46 |
10-Leaf |
35 |
31 |
38 |
25 |
40 |
40 |
11-Twig |
35 |
24 |
45 |
27 |
30 |
44 |
12-Leaf |
40 |
22 |
35 |
30 |
35 |
34 |
13-Twig |
38 |
22 |
32 |
25 |
-- |
41 |
14-Leaf |
40 |
30 |
39 |
26 |
35 |
-- |
15-Twig |
35 |
30 |
31 |
26 |
51 |
40 |
16-Leaf |
47 |
24 |
34 |
29 |
46 |
30 |
17-Twig |
39 |
35 |
35 |
31 |
37 |
38 |
18-Leaf |
55 |
22 |
34 |
30 |
42 |
39 |
19-Leaf |
41 |
29 |
35 |
58 |
50 |
50 |
20-Leaf |
48 |
26 |
30 |
22 |
52 |
65 |
21-Leaf |
34 |
30 |
32 |
27 |
-- |
29 |
22-Leaf |
59 |
24 |
40 |
38 |
58 |
19 |
23-Leaf |
37 |
23 |
40 |
30 |
31 |
46 |
24-Twig |
43 |
35 |
30 |
25 |
35 |
34 |
Table 3: Highest zones of inhibition observed at day 4 for well diffusion anti-microbial assays of extracts from 24 bacterial endophytes extracted using different solvents. Zones were measured at 50 ul of extracts with S. aureus and E. coli being used as tester strain.
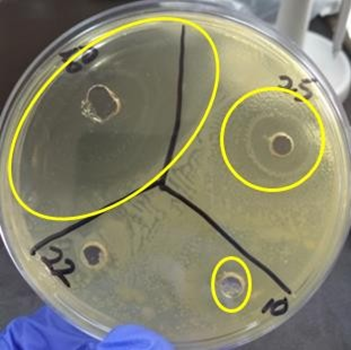
Figure 1: Zone of inhibition of the most potent bioactive endophytic extract 22 viewed after 72 hours of incubation at 37oC against S. aureus.
Antibacterial assay with ampicillin as a controlAfter testing the effect of extracts from 24 endophytes against E. coli and S. aureus antimicrobial activity was noted and secondary assay was done. For the secondary assay, acetone extracts were plated on the same plate together with ampicillin in order to compare potency against S. aureus. The secondary disc diffusion assay was used where effect of 10 and 50 ul of extracts and 10 ul of ampicillin were measured. Zones of inhibition of ampicillin were ranging from 23 to 36 mm. For the 24 samples volumes of 10 ul gave zones of inhibition ranging from 26 mm to 38 mm whilst 50 ul gave 35 mm to 58 mm. zones of inhibition were noted after 48 hours (Table 4).
| Sample | Ampicillin | Extract | |
|---|---|---|---|
10 ul Inhibition zones/mm |
10 ul Inhibition zones/mm |
50ul Inhibition zones/mm |
|
1-Leaf |
26 |
27 |
54 |
2-Leaf |
26 |
30 |
50 |
3-Twig |
36 |
38 |
45 |
4-Leaf |
27 |
26 |
35 |
5-Leaf |
31 |
31 |
38 |
6-Twig |
27 |
33 |
44 |
7-Twig |
29 |
34 |
49 |
8-Twig |
32 |
30 |
48 |
9-Leaf |
31 |
35 |
45 |
10-Leaf |
31 |
30 |
51 |
11-Twig |
25 |
31 |
51 |
12-Leaf |
30 |
31 |
46 |
13-Twig |
25 |
30 |
38 |
14-Leaf |
27 |
34 |
46 |
15-Twig |
25 |
29 |
49 |
16-Leaf |
36 |
30 |
44 |
17-Twig |
32 |
27 |
43 |
18-Leaf |
30 |
29 |
49 |
19-Leaf |
26 |
32 |
58 |
20-Leaf |
28 |
30 |
47 |
21-Leaf |
33 |
32 |
40 |
22-Leaf |
29 |
32 |
45 |
23-Leaf |
26 |
32 |
40 |
24-Twig |
29 |
32 |
40 |
Table 4: Relative sizes of zones of inhibition obtained after incubating tester strain (S. aureus) with 24 different endophytes obtained from twigs and leaves of mutowa tree and ampicillin (control) for 48 hours.
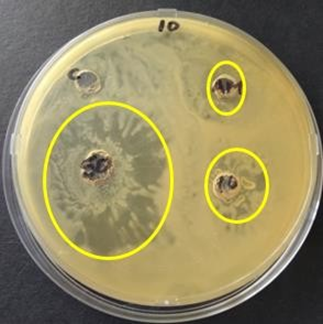
Figure 2: Zones of inhibition of endophytic extracts in comparison to ampicillin (AM) of concentration 16mg/ml observed after 48 hours of incubation at 37oC against S. aureus.
16S ribosomal RNAAmplification of 16S rRNA gene was successful in all twenty-four samples as shown in figure 3. This indicated that all endophytes were prokaryotic in nature. 27F and 1492R primers were used because they can bind to highly conserved regions within the 16S rRNA gene, present in all prokaryotic cell [15].
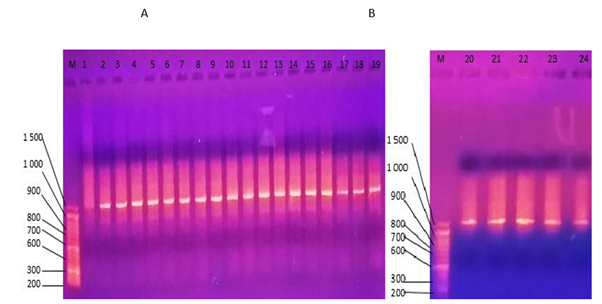
Figure 3: A and B showing ethidium bromide stained gel electrophoresis of 16S rRNA gene polymerase chain reaction from extracted genomic DNA.
DNA sequence analysisSequencing of 16S rRNA gene amplicons was delayed due to the Covid 19 pandemic inconveniences therefore only isolates 5, 7, 8, 9, 15, 16, 22, 23 and 24 were sequenced. These isolates were selected basing on those that gave higher zones of inhibition on antimicrobial assays. BLAST was used to reveal sequence identities which are summarized in table 5. Sequencing of the 16S rRNA gene identified the isolates to strain level.
| Isolate | Identity | Percentage identity | Accession Number |
|---|---|---|---|
5- Leaf |
Izhakiella capsodis |
96% |
NR_148767.1 |
7- Twig |
Escherichia fergusonii |
96% |
NR_074902.1 |
8- Twig |
Izhakiella capsodis |
95% |
NR_148767.1 |
9- Twig |
Escherichia fergusonii |
96% |
NR_114079.1 |
15- leaf |
Shigella dysenteriae |
93% |
NR_026332.1 |
16- Twig |
Xenorhabdus szentirmaii |
93% |
NR_042328.1 |
22- Leaf |
Providencia rettgeri |
87% |
NR_042415.1 |
23- Leaf |
Dickeya zeae |
96% |
NR_041923.1 |
24- Leaf |
Escherichia albertii |
96% |
NR_025569.1 |
Table 5: Species identities.
Phylogenetic analysisThe nine sequences were used to generate a phylogenetic tree (Figure 4). Three notable branches were observed from the tree. Escherichia albertii had no recent relative to any of the sequences other than the ancestral root. This indicates that Escherichia albertii diverged early and has a few changes from the most ancestral stain [10]. Shigella dysenteriae, Xernorhabdus szentirmaii, Providencia rettgeri and Dickeya zeae emerged from a common ancestor indicating that they are closely related. However, Providencia rettgeri and Dickeya zeae share most recent ancestor making them more closely related species and recently diverged species in that cluster. Location of two strains of Escherichia fergusonii indicated that they diverged at the same time which is expected as they are same strains. Phylogenetic analysis indicated that two species of Izhakiella capsodis diverged at different times. Full genome sequencing would have given more insight on genetic differences between the two Izhakiella capsodis strains.
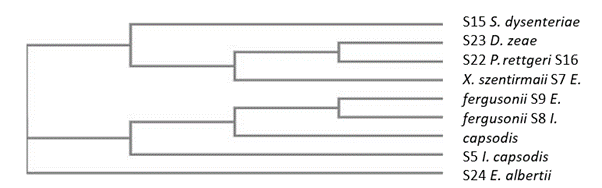
Figure 4: Phylogenetic tree constructed with sequences of the 16S rRNA regions of endophytic bacteria isolated from mutowa tree.
Restriction mappingThe link https://www.takara-bio.co.jp/enzyme/enzyme_search.php was used to develop restriction maps for two sequences of Izhakiella capsodis. This was done to compare similarity and differences of the 16S rRNA gene of the two sequences. From the results obtained using restriction enzyme EcoRI and Hind III it was observed that the sequences belonged to two different species of Izhakiella capsodis as shown in figure 5.
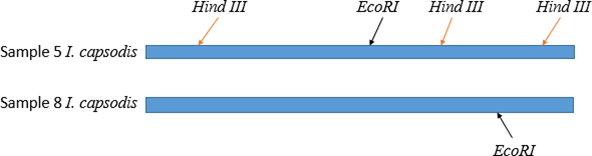
Figure 5: Restriction sites of restriction enzymes (Hind III and EcoRI) on the 16S rRNA gene sequence of two Izhakiella capsodis species (sample 5 and 8).
Pathway mappingPathway maps of sequenced isolates were obtained using Kyoto Encyclopedia of Genes and Genomes (KEGG) database. From the database only pathway maps for Shigella dysenteriae, Providencia rettgeri, Dickeya zeae, Escherichia albertii and Escherichia fergusonii were found. Pathway maps for Izhakiella capsodis and Xenorhabdus szentirmaii were not found on the database. Pathway maps showed that isolates have potential to biosynthesize antibiotic compounds shown in table 6. KEGG pathway mapping results were in agreement with the antimicrobial assays findings.
| Species | Bioactive compounds | Function |
|---|---|---|
(Escherichia fergusonii, Escherichia albertii and Shigella dysenteriae) |
Streptomycin, Monobactam, Novobiocin Carbapenem |
Highly effective antibiotics, used to treat serious bacterial infections Beta lactam highly effective antibiotic used to treat high risk bacterial infections. Therapeutic drug monitoring important |
Acarbose |
Slows digestion of carbohydrates, helps low blood sugar levels. It is used treat type 2 diabetes |
|
Validamycin |
Aminoglycoside antibiotic and fungicide |
|
Dickeya zeae |
Streptomycin, Monobactam Novobiocin, Roseoflavin and Carbapenem Acarbose Validamycin |
Highly effective antibiotics Highly effective antibiotics Used to treat type 2 diabetes Fungicide and antibiotic |
Cycloserine |
Used to treat active drug resistant tuberculosis |
|
Providencia rettgeri |
Streptomycin, Monobactam, Bacilysin, Aurachin, Novobiocin, Kanosamine, Fosfomycin Acarbose Validamycin, Capsaicin Podopyllotoxin Puromycin Fumiquinazoline D |
Highly effective antibiotics Highly effective antibiotics Used to treat type 2 diabetes Aminoglycoside antibiotic and fungicide Used to treat arthritis, backache and as an analgesic Used to treat genital warts and molluscum cotagiosum Aminonucleoside antibiotic, inhibits protein synthesis Antibiotic activity against gram positive and gram negative |
Table 6: Predicted bioactive compounds synthesized by sequenced strains from Diplorhynchus condylocarpon using KEGG.
The 16S rRNA gene is a useful molecular marker that can be used to identify species. Endophytes from Diplorhynchus condylocarpon are capable of synthesizing and are reservoirs of bioactive compounds. Some of these bioactive compounds are the ones that made it possible to use extracts from Diplorhynchus condylocarpon in antimicrobial assays and should be explored further. These endophytes produce highly effective antibiotics that uses numerous mechanisms to attack both Gram-negative and Gram-positive bacteria. Bioinformatics tools such as KEGG are useful in analyzing genes present in a species. Through KEGG not only antibiotic biosynthesis genes were identified but also compounds such as Acarbose which can be used in the treatment of diabetes. Ability of endophytes to produce antibiotics and other bioactive compounds can be exploited for pharmaceutical use.
We acknowledge the technical assistance of Mr Hillary.
Citation: Ntobeko Dube and Idah Sithole-Niang. “Molecular Characterisation of Bacterial endophytes from the Medicinal Plant Diplorhynchus condylocarpon". Acta Scientific Paediatrics 5.3 (2021): 82-90.
Copyright: © 2021 Ntobeko Dube and Idah Sithole-Niang. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.