Kulvinder Kochar Kaur1*, Gautam Nand Allahbadia2 and Mandeep Singh3
1Scientific Director Cum Owner, Dr Kulvinder Kaur Centre for Human Reproduction Scientific Director Cum Owner, Punjab, India
2Scientific Director, Ex-Rotunda-A Centre for Human Reproduction, Bandra, Mumbai, India
3Neurology, Consultant Neurologist, Swami Satyanand Hospital, Baradri, Jalandhar, Punjab, India
*Corresponding Author: Kulvinder Kochar Kaur, Scientific Director Cum Owner Dr Kulvinder Kaur Centre for Human Reproduction Scientific Director Cum Owner, Punjab, India.
Received: September 11, 2024; Published: September 24, 2024
Citation: Kulvinder Kochar Kaur., et al. “Potential Targeting of Differentially Expressed Genes Regarding CAR -T -Cells in Ovarian Cancer (OC) Treatments: A Future Prospective Advancement for Improvement of 5 yr Survival-A Short Communication”. Acta Scientific Cancer Biology 8.10 (2024): 02-06.
Recently we had reviewed the commonest of Epithelial ovarian cancer (EOC’s), namely high grade serous ovarian cancers (HGSOC)-70% of all EOC’s, manner to improve 5 yr survival rate in the HGSOC, with emphasis on Homologous Recombination Deficiency and Intra tumor heterogeneity, however in view of late diagnosis of EOC’s generally incomplete debulking surgery done as well as generation of resistance to platinum dependent chemotherapy. In view of resistance to chemotherapeutic agents might takes place buttresses the urgent mandatory requirement for the innovative therapeutic approaches Here we further present how chimeric antigen receptor (CAR)-T cell treatments might become future targets following bioinformatics study where isolation of 9 proteins including MUC1, CXCR4, EpCAM, RACGAP1, UBE2C, PRAME, SORT1, JUP as well as CLDN3 in the form of crucial proteins in the oncogenic pathways of OC, therefore marking them being significant antigen candidates regarding interventions for CAR-T-cell. These proteins, on the plasma membrane as well as anticipated to be oncogenic, yield a substantially greater magnitude of specificity for plausible targeted therapies. Nevertheless, in view of the baseline evaluation was computational, extra experimental as well as clinical corroborations are imperative to validate such preliminary observations along with accurately perform analysis of the effectiveness of such proteins in the form of CAR-T- cell antigens. This offers an innovative exciting immunometabolic leap in therapy of OC’s for overcoming the problem of chemotherapy resistance,furtherescalating5 yr survival rate in all OC’s .
Keywords:Cancers; Chimeric Antigen Receptor (CAR); T Cells
Amongst Ovarian Cancers, Epithelial ovarian cancer (EOC) possesses the maximum prevalence. EOC’s display different kinds of histopathological, immunohistochemical, as well as genomic characteristics out of which 5 primary kinds have been isolated: i) High Grade Serous Ovarian Cancer (HGSOC) (70%), ii) endometrioid, carcinomas (10%) iii) Clear cell carcinomas (10%) iv) Mucinous carcinomas (3%) I as well as v) Low grade serous Ovarian Cancer (LGSOC) (<5%) [1]. Typically, the presentation of these HGSOC is in considerably advanced International Federation of Gynecology and Obstetrics (FIGO) stages of the disease- for instance FIGO stage III-IV in view of its asymptomatic presentation which makes it imperative for the performance of primary debulking surgery followed by adjuvant chemotherapy or neoadjuvant chemotherapy (NACT) with the interval surgery. Reason for the manifestation of OC diagnosis gets established only in substantially advanced disease is that there is no partition amongst abdominal organs and pelvis [2].
The generation of a chimeric receptor for T-cells was done by Eshhar Z in 1993 with the idea of escalating efficacy in addition to plausibility of generating efficacious anti tumor treatment [3]. Of the maximal impacting along with particular methodologies of immune treatment is portrayed by chimeric antigen receptor (CAR)-T-cell treatments. CAR -T cells treatments have come up in the form of leading advancement, that result in considerable plausibility of converting cancer therapy approaches [4]. CAR -T cells treatments have attained attractive status in the form of an immunometabolism approach for the management of variety of malignancies. Fashioning of such engineered T cells is performed in a manner that it possesses the capacity of recognition in addition to breakdown of the cancer cells which are expressing particular antigen overtaking the major histocompatibility complex (MHC) limitations correlated with canonical T cell activation. Although, success has been attained by CAR -T cell treatments in reference to haematological malignancies, its plausibility in solid tumors for instance ovarian cancer (OC) has not been totally estimated. Crucial hurdles are inclusive of the isolation of the antigen which have remarkable expression in tumor cells, however are practically negligible existent in normal tissues.
OC’s continue to be major hurdle in reference to gynaecological oncolology as well as is implicated significantly in causation of mortality along with morbidity for the women globally. In maximal patients with manifestation of OC, diagnosis gets established only in substantially advanced disease stage in view of non-particular symptomatology along with absence of efficacious determining methodologies. The prevalence varies amongst 9.2-12.0/10,000 women all over world [5]. This is akin to the case presented by us recently in a 65 yr old post-menopausal lady of HGSOC that simulated a twisted ovarian cyst in a completely asymptomatic healthy lady [6]. Despite the median age regarding diagnosis differs amongst 50-79 yrs, an elderly age escalates the risk of greater hostile kinds of OC [7]. Canonical approaches for the management for instance surgery or platinum dependent chemotherapy, yielded restricted enhancement of the 5yr overall (OS) survival rates as well as induction of resistance to chemotherapeutic agents might takes place [8], which buttresses the pacey mandatory requirement for innovative therapeutic approaches. Genetic engineering of immune working cells whose programming had been done with the idea of recognition of particular antigen, give a strategy which might prove to be path breaking strategy meant for targeting in addition to depleting malignant cells in a selective manner. Isolation of target antigens, specifically for particular cancer kinds of antigen, continue to be major challenge, however it is crucial in reference to successful CAR -T cells treatments [9]. The anticipated mechanistic modes are emphasized in figure 1.
Recently we had detailed natural killer cells (uNK cells) kinds of killer immunoglobulin-like receptors (KIR), whose binding takes place with their respective HLA ligands in reference to recurrent implantation failure (RIF) uNK cells activation takes place as well as liberation of cytokines which facilitates placentation. The 16 are considerably polymorphic [7], along with might be grouped in the form of “activating” or ‘’hampering’’. Binding of these activating KIRs with their HLA ligands result in activation of the NK cells along with cytotoxicity, while that with the hampering KIR causes repression of NK cells working (reviewed in ref no 10). Same works in CAR-NK cells killing mode [10]. Readers are referred to reference 10 for detailed modes why CAR - NK cells are preferred.
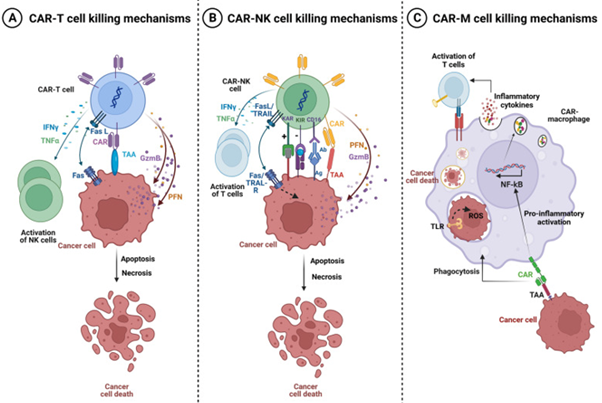
Figure 1: Courtesy ref no-9-Killing mechanisms of CAR-T, CAR-NK, and CAR-M cells. A Tumor killing mechanisms of CAR-T cells. Activated CAR-T cells can specifically recognize the tumor associated antigen (TAA). Cytotoxic activity of Chimeric Antigen Receptor (CAR)-T cells is mediated by perforin (PFN) and granzyme (GzmB) granules secretion, and by activation of death receptor pathways such as Fas/Fas-L leading to cancer cells apoptosis and necrosis. Activated CAR-T cells also secrete Interferon-gamma (IFN-γ) and tumor necrosis factor-alpha (TNFα) which can promote Natural Killer (NK) cell anti-tumor cytotoxic activity. B Tumor killing mechanisms of CAR-NK cells. The activity of CAR-NK cells is regulated by the signal of activating (KAR) and inhibitory receptors (KIR) expressed on NK cells. Activated CAR-NK cells secrete the cytotoxic proteins perforin and granzyme B which synergize to induce cancer cell necrosis and apoptosis. NK cells also express the death ligands FasL and TRAIL which will bind to Fas and TRAIL-R on cancer cells and induce apoptosis. Moreover, CAR-NK cells trigger ADCC through the CD16 Fc receptor which recognize antibody-opsonized cancer cells. In addition, CAR-NK cells secrete IFN-γ and TNFα which promote their activation and stimulate other T-lymphocytes leading to increased anti-tumor immune response. NK: cell-Natural killer cells; IFN-γ: Interferon-gamma; TNFα: Tumor necrosis factor-alpha; TRAIL-R: TNF-related apoptosis-inducing ligand, KIR: Killer Inhibitory Receptors, KAR: Killer Activation Receptor, ADCC: Antibody-dependent cellular cytotoxicity, (PFN) perforin and (GzmB) granzyme. C Tumor killing mechanisms of CAR-M. The binding of a specific tumor associated antigen (TAA) with CAR receptor on the surface of CAR-M generates activation signals that mediate tumor phagocytosis, activation of transcription factors such as NF-kB and subsequent release of pro-inflammatory cytokines, which in turn can activate T cell-mediated immunity against the tumor.
The plausibility of utilizing of CAR-T-cell treatments in case of OC is escalating getting recognition, nevertheless our insight regarding appropriateness of antigen targets continues to be in substantially early state. Giving significance to tackle existence of knowledge gap for generation of greater efficacious, individualized CAR-T-cells treatments in case of OC is the urgent need. In this decade of advancements regarding precision medicine, one is not exaggerating over part played by bioinformatics. Bioinformatics portrays utilizing computational gadgets for management, evaluation in addition to interpretation of the biological outcomes, has assumed major position in reference to cancer research, specifically in this decade of high- throughput genomics. Incorporation of the genomic, transcriptomic, proteomic outcomes from several resources might yield exhaustive panorama of tumor biology, promoting the invention of plausible targets for innovative therapeutics [11]. Recently Anurogo., et al. [12], with the goal of isolation of plausible genes in the form of CAR -T -cells antigen candidates regarding OC They conducted assessment of differentially expressed genes (DGE) on ovarian cancer samples from four datasets derived from the gene expression omnibus (GEO) datasets. Working elucidation by pathway evaluation, protein placement in addition to gene expression evaluation was performed with utilization of the separate datasets as well as gadgets. An assessment of oncogenicity along with network evaluation was further conducted. Totally 153 DGE were isolated in OC samples, with expression of 60 DGE expressing plasma membrane (PM)protein appropriate for the CAR -T -cells antigens. Of these 21 PM proteins were anticipated to be oncogenes in ovarian cancers, with 9 proteins having key part in the network. Crucial genes isolated in the oncogenic pathway in case of OC were inclusive of transmembrane glycoprotein mucin 1 (MUC1), The C-X-C Motif Chemokine Receptor 4 (CXCR4), epithelial cell adhesion molecule protein (EpCAM), Rac GTPase activating protein 1 (RACGAP1), ubiquitin-conjugating enzyme E2C (UBE2C), Preferentially Expressed Antigen in Melanoma protein (PRAME), Sortilin 1 (SORT1), Junction Plakoglobin, also known as plakoglobin (JUP), claudin 3 protein (CLDN3) pointing these in the form of advocated antigens in reference to CAR-T-cells treatments [12]. (Figures 2-4).
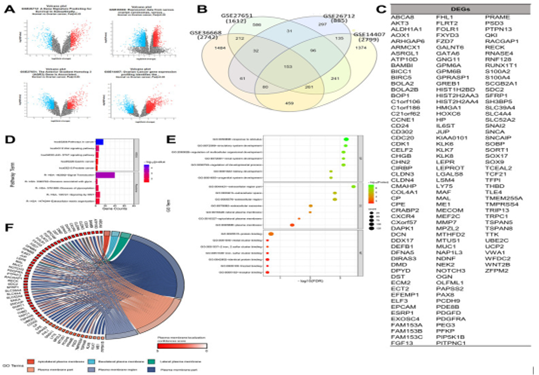
Figure 2: Courtesy ref no-12-Determination of gene set enrichments (GSEs) and differentially expressed genes (DEGs). (A) Volcano plot of gene distributions in control and ovarian cancer samples. Gray dots represent genes that are not significantly expressed between normal and ovarian cancer cell samples. (B) Venn diagram of overlapping gene between four GSEs from which we obtained 153 DEGs. Also, the highest unique genes are GSE36668, GSE14407, and GSE27651. (C) List of DEGs. (D) Pathway terms related to DEGs, colored by -log10 (p-values). (E) Gene ontology terms related to DEGs, colored by –log10 (p-values). (F) Plasma membrane-related genes from six GO terms (p > 0.05) with localization confidence scores. A higher score means a greater probability that the protein will be situated ther.
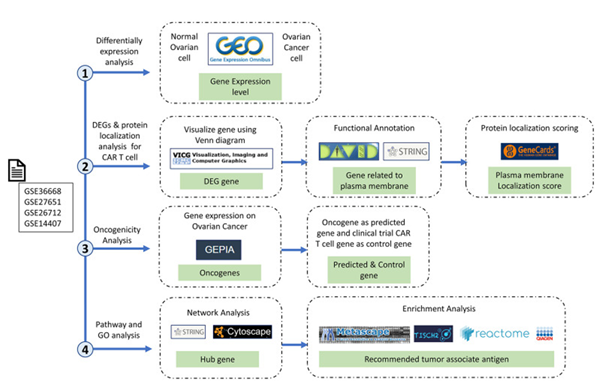
Figure3: Courtesy ref no-12-Research stages and workflow. The four research red lines are differential expression analysis, differentially expressed genes (DEGs), protein localization analysis, oncogenicity analysis, and pathway and gene ontology analyses. Therefore, in this study, we carried out multilevel screening to reduce the potential for errors or discrepancies later. What we mean by multilevel screening is first looking at the significance of the expression of a gene in normal and cancer samples from the dataset, then checking the significance of the significant gene again in a different database to ensure that the gene is significantly expressed only in cancer cells.
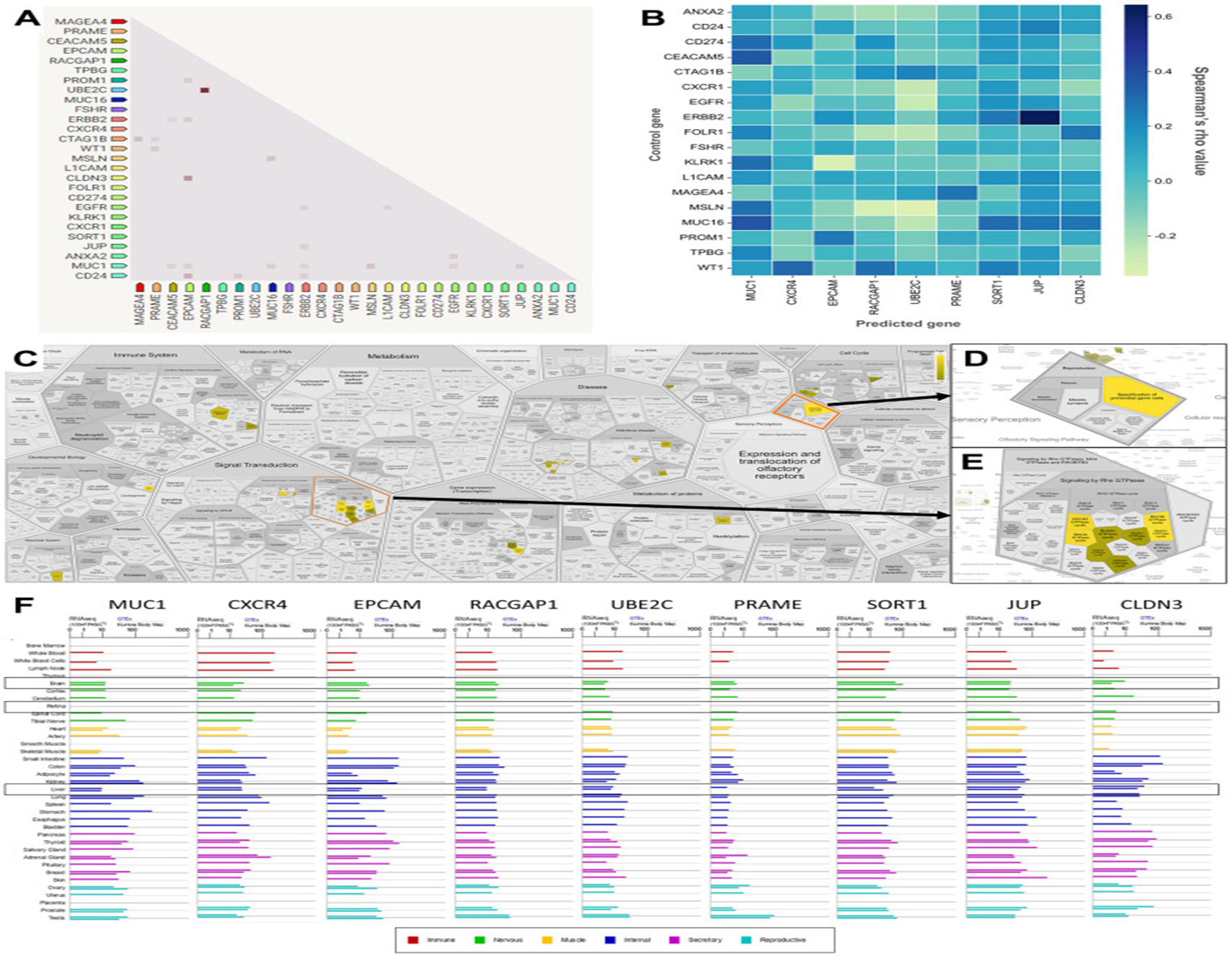
Figure 4: Courtesy ref no-12-Correlation, co-expression, and pathway analysis of TAAr. (A) Co-expressed genes are colored by a dot; a darker dot means a higher co-expression score. Co-expression score: the higher the score, the higher the probability that co-expression will occur. (B) Spearman’s correlation scores for gene expressions in ovarian cancer (OC), which are comprised of 27 proteins consisting of nine TAAr and 18 control genes. (C) The complete pathway of the Reac tome database related to TAAr. This pathway was constructed with the Voronoi tessellation method termed Reac Foam, which provides user-friendly access and visualization. (D) Pathway related to reproduction where the specification of primordial germ cells pathway is located. (E) Rho GTPase-related and neighbour pathways. (F) The expression data of TAAr in brain, retina, and liver tissue, provided by GeneCard, were used to analyze the possible toxic effects of targeting TAAr.
CAR T cell treatment has illustrated promise in treating hematological malignancies, with the plausibility of maintainance of remissions in addition to improvement of clinical result [13]. This is in view of the healing efficacy of CAR therapy in acute lymphocytic leukemia patients, which might be escalated upto 92%. CARs function by targeting the tumor associated antigen (TAA) observed on the cell surface as well as bypassing the event of recognition with MHC class I or II [14]. Nevertheless, the generation of along with research into CARs in solid tumors are still restricted inclusive of in OC patients. OC is a kind of cancer in addition to an orphan drug disease with a remarkably greater mortality rate in view of malignancies [15].
In this research, the topography of plausible antigenic targets for CAR-T-cell therapies in OC has been significantly widened. Application of bioinformatics procedures for the assessment of DEGs as well as protein-protein interactions (PPIs), the study has emphasized on nine proteins (MUC1, CXCR4, EPCAM, RACGAP1, UBE2C, PRAME, SORT1, JUP, and CLDN3) in the form of crucial proteins in the oncogenic pathways of OC, therefore marking them being significant antigen candidates regarding interventions for CAR-T-cell. These proteins, on the plasma membrane as well as anticipated to be oncogenic, yield a substantially greater magnitude of specificity for plausible targeted therapies. Nevertheless, in view of the baseline evaluation was computational, extra experimental as well as clinical corroborations are imperative to validate such preliminary observations along with accurately perform analysis of the effectiveness of such proteins in the form of CAR-T- cell antigens. Thereby, this study despite certain caveats, mirrors a significant jump in the ongoing research for detailed in addition to rigorous immunotherapies for OC.
Copyright: © 2024 Kulvinder Kochar Kaur.,et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.